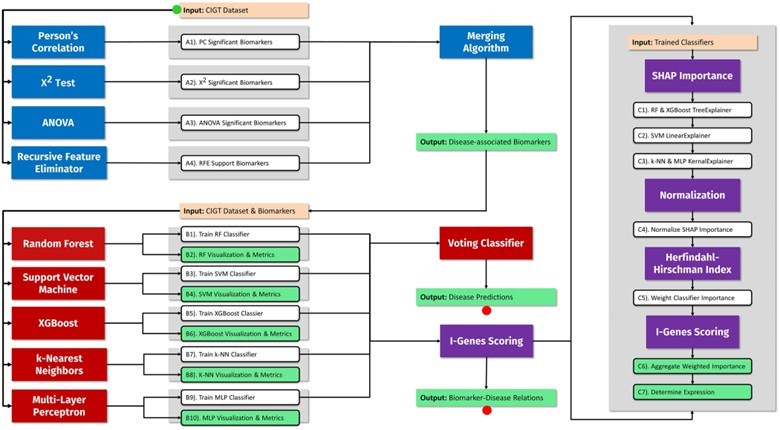
A pipeline combining classical statistics and machine learning algorithms is used to identify disease-associated biomarkers and predict disease risk
Invention Summary:
Gene Expression Analysis and Genome‐wide association studies provide an unprecedented understanding of the genetic basis of human disease by uncovering millions of loci associated with various complex disease phenotypes. However, these are unable to completely map biomarkers and predict disease with high accuracy. The key limitation is our lack of technology to analyze the complete genome or transcriptome of patients to identify all the genetic components of complex traits.
To address this, researchers at Rutgers University have developed a software pipeline to help predict diseases in individuals by combining conventional statistical methods with cutting-edge machine learning algorithms to measure the significance of genomic biomarkers. The pipeline can intake data including patient demographics, genomic and transcriptomic data to produce personalized patient predictions and visual representations of the biomarkers significant to disease prediction.
Market Applications:
-
Application integrated with Electronic Health Records (EHR) systems
-
Precision Medicine, personalized genetic insights, sequencing as a service companies
-
Research institutions / universities
-
Diagnostic Lab Testing
-
Pharmaceutical companies
Advantages:
-
Comprehensive biomarker discovery
-
Two-in-one functionality: identifies disease-associated biomarkers and predicts patient diagnoses
-
Easy to use interface, allowing those without prior AI background to use the platform
-
Does not require access to high-performing computers
-
Market for application of AI to precision medicine is ~ USD 1.3 billion in 2022
Intellectual Property & Development Status: Patent pending. Software available for licensing and/or research collaboration. For any business development and other collaborative partnerships contact marketingbd@research.rutgers.edu.
Publication available: https://academic.oup.com/bioinformatics/article/39/12/btad755/7473370