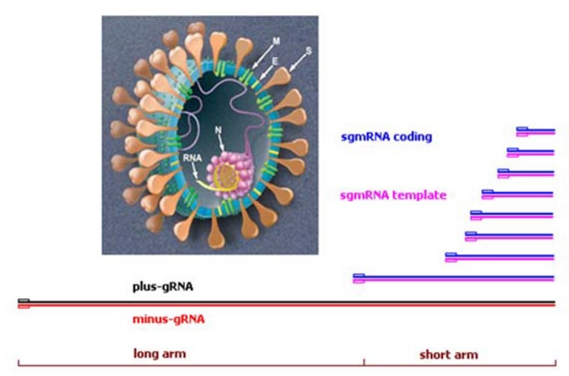

Schematics of SARS-CoV-2 genome and subgenomic mRNAs. Production of sgmRNAs can introduce unusual deletions and junctions in COVID variants.
Invention Summary:
Although COVID-19 emergency is “officially over”, the potential threat of novel variants, which may lead to severe disease, has not disappeared. Despite the huge numbers of publications on SARS-CoV-2 in the past 4 years, the research community has only scratched the surface of many aspects of virus biology. For example, analysis of patient samples shows that novel open reading frames and genes appear in variants of concern that have not been studied in depth. These genetic variations may lead to novel, unknown mRNAs and proteins produced by the virus and are specific for different variants of concern.
Rutgers researchers have identified several such non-canonical variations of viral RNA in patient samples. This discovery can be utilized to predict if a new virus variant in a patient is more Omicron‐like, and thus likely more mild and less pathogenic. A set of PCR primers can be used to search for this specific subgenomic mRNAs in a patient sample.
Market Applications:
- COVID-19 diagnostic tests, including a PCR test that can detect non-canonical RNA variants and inform disease severity
- Analogous tools for other coronaviruses
Advantages:
- Prediction of analogous events in patient samples for known and unknown coronaviruses (e.g., SARS, MERS, COVID or novel diseases)
- Characterization of patient COVID variants based on severity
- Public health awareness regarding development of new strains
- Furthering our understanding of viral biology
Intellectual Property & Development Status: Patent pending. Software available for licensing and/or research collaboration. Available for licensing and/or research collaboration. For any business development and other collaborative partnerships contact: marketingbd@research.rutgers.edu