
Invention Summary:
The CRISPR/Cas9 system is a powerful tool for targeted genetic engineering in
organisms ranging from animals to plants. Cas9 has intrinsic nuclease activity, which causes sequence specific DNA double-strand breaks (DSBs), leading to
activation of cellular pathways such as homology dependent repair (HDR) for gene conversion and nonhomologous end joining (NHEJ) for gene inactivation.
However, the dependence on DSBs may lead to undesired genetic events such as off-target insertions, deletions, and chromosomal translocations.
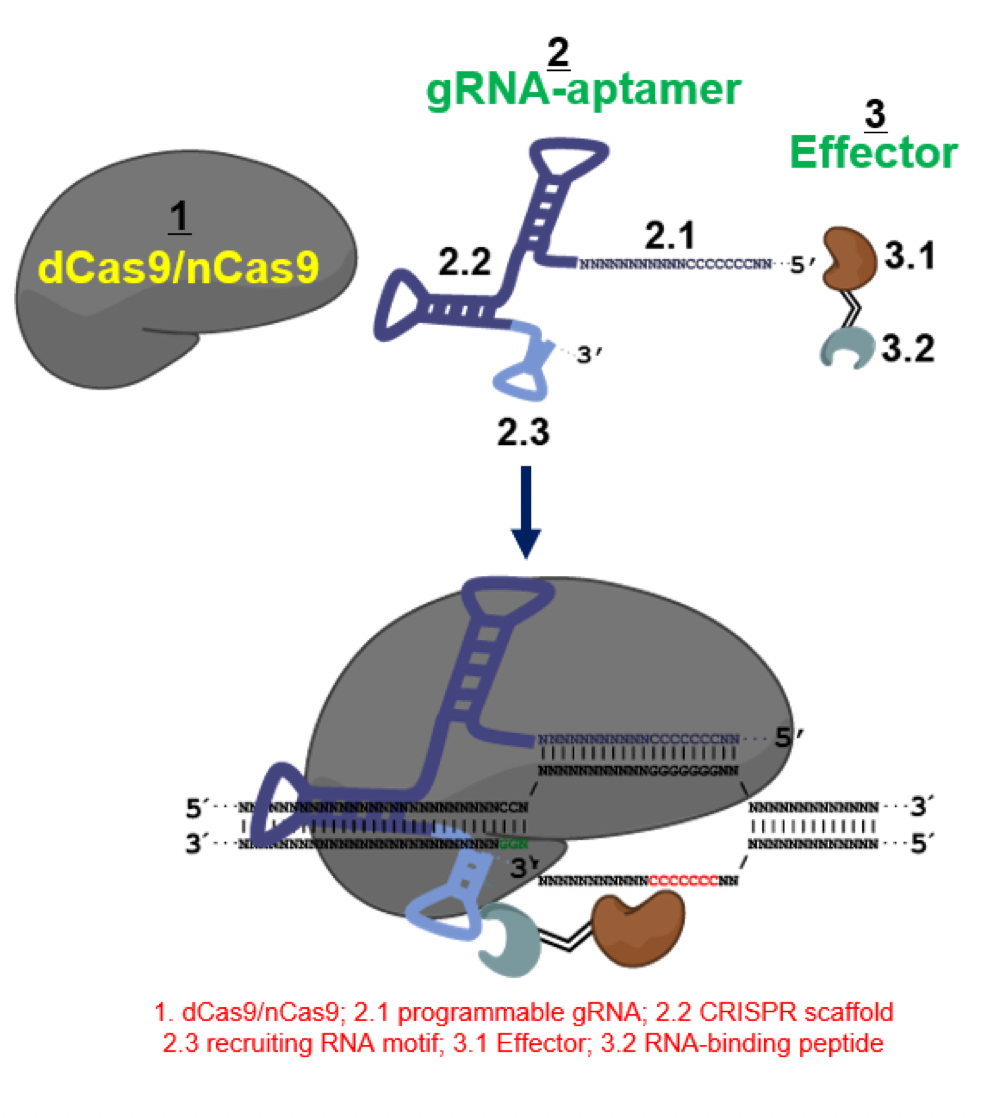
The Base Editing technology overcomes the requirement of DNA DSBs for gene editing. Rutgers researchers have developed one of the two base editing platform technologies that allow sequence directed base editing in DNA or RNA. The system features: (1) a modular design, (2) high efficiency, (3) low indel rates, (4) low to absent of off-target effect, (5) activity independent of DNA DSBs or HDR.
The system contains three modules as illustrated above: (1) a nickase CRISPR/Cas9 module; (2) an RNA scaffold module for sequence targeting and for
recruitment of a gene conversion effector; and (3) an effector module with DNA or RNA modifying enzyme activity.
Market Applications:
• Therapeutics for genetic diseases in particular those caused by certain point mutations*
• Genetic engineering in animals*
• Plant genetic engineering
• Research tools
*: Animal and therapeutic application rights have been licensed.
Intellectual Property & Development Status: Licensing opportunities available for AGRICULTURE use only.
• Independent base editing platform
• World-wide composition and method protection. PCT/US2016/042413
• Priority date: July 15, 2015
• Patent allowed in the European Union
Additional reference:
• Juan, C. et al. CRISPR Journal. doi.org/10.1089/crispr.2020.0035.