SuperSelective PCR Primers
 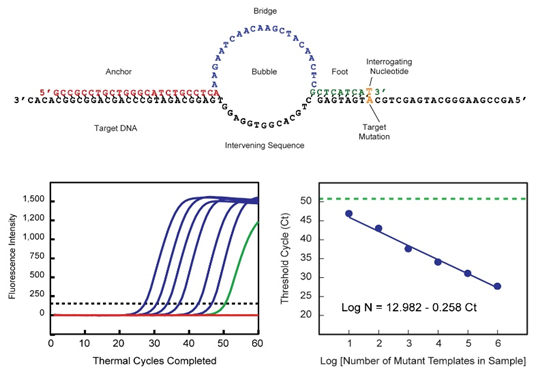
Structure of a SuperSelective primer for the detection and quantitation of rare mutant sequences in the presence of 1,000,000 wild-type sequences; and a demonstration that the number of amplification cycles required to generate a signal above background is inversely linearly proportional to the logarithm of the number of mutant molecules in the original sample.
Invention Summary:
“SuperSelective” primers enable the detection and quantitation of somatic mutations whose presence relates to cancer diagnosis, prognosis, and therapy in real-time PCR assays that can analyze rare DNA fragments present in blood samples (or in other biological tissues).
Rutgers researchers have created a design of these deoxyribonucleotide primers which includes both a “5' anchor sequence” that hybridizes strongly to target DNA fragments and a short, physically and functionally separate, “3' foot sequence” that is perfectly complementary to the mutant target but mismatches the wild type. As a consequence, amplification mainly occurs on the mutant target sequences. As shown in the figure above, as few as ten mutant fragments can be detected in the presence of 1,000,000 wild-type fragments, even when the difference between the mutant and the wild-type is only a single nucleotide polymorphism. Multiplex PCR assays employing a set of SuperSelective primers can utilize differently colored molecular beacon probes or sloppy molecular beacon probes, each specific for the amplicons generated from a different target mutation, enabling a comprehensive assessment of the relevant somatic mutations present in a patient's clinical sample.
Market Applications:
- Identification and quantification of somatic mutations indicative of cancer diagnosis, choice of therapy, and early detection of recurrence, utilizing non-invasive liquid biopsies (or tissue samples)
- Identification of rare antibiotic resistance mutations in bacterial and fungal samples
- Potential to detect cancer in a routine blood sample taken during an annual medical examination before any symptoms have occurred
Advantages:
- Assays are carried out on widely available spectrofluorometric thermal cyclers
utilizing commercially available buffers and take only a few hours to complete
- Closely related sequences are much less likely to be amplified
- Sensitivity is greater than analyses utilizing more expensive next-generation sequencing
- SuperSelective PCR primers enable a large number of somatic mutations to be simultaneously identified
Intellectual Property & Development Status:
- Highly Selective Nucleic Acid Amplification Primers – US Patents 9,909,159;
US 10,815,512; 11,111,515 and related foreign patents.
- Multiplex Nucleic Acid Assays Capable of Detecting Closely Related Alleles and Reagents Therefore – US Patent 11,542,547 and related foreign patents.
- Assay Methods and Kits for Detecting Rare Sequence Variants – Pending US Patent Application 17/754,032 and related foreign patents.
- Use of Structure-Specific Amplification Primers for the Detection of Genetic Rearrangements – Nonprovisional US Patent Application will be submitted in 2024.
Additional publications and patent applications:
For any business development and other collaborative partnerships, contact: marketingbd@research.rutgers.edu
Patent Information:
| Title |
App Type |
Country |
Serial No. |
Patent No. |
File Date |
Issued Date |
Expire Date |
Patent Status |
|
|
|
ID: S2016-121
Category:
Inventors:
Keywords:
|